Bibliometrix package
By Tainá Rocha based on Beatriz Mils post https://beatrizmilz.com/blog/
11/12/2021
A package for quantitative research in scientometrics and bibliometrics that includes all the main bibliometric methods of analysis.
1. Get the dataset
I obtained the data from SCOPUS and Web of Science repositories (WOS). I searched for articles using a generic query for test purposes. It is important to emphasize that building a correct query requires some steps
Generic query just to test package: Reforestation * Temperate forest
Years: from 2000 to 2021
Finally, select all items of search to export. In this tutorial I exported as “.bib” file
SCOPUS
WOS
2. The package
2.1 To install
#install.packages("bibliometrix")
2.2 Shiny
The package presents a Shiny version, which makes it possible to run analyses in a point and click interface, without write code. This is a really cool possibility for people who want to use the tool but don’t want to write code in R. To use biblioshiny, you need to run the below code without # :
# bibliometrix::biblioshiny()
2.3 Despite Shiny it’s really cool, writing the code it’s a better way for reproducibility in future analyses. Also, It’s easy to use codes for this package.
Loading some libraries (make sure that all libraries are installed)
library(dplyr)
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
library(bibliometrix)
## To cite bibliometrix in publications, please use:
##
## Aria, M. & Cuccurullo, C. (2017) bibliometrix: An R-tool for comprehensive science mapping analysis,
## Journal of Informetrics, 11(4), pp 959-975, Elsevier.
##
##
## https://www.bibliometrix.org
##
##
## For information and bug reports:
## - Send an email to info@bibliometrix.org
## - Write a post on https://github.com/massimoaria/bibliometrix/issues
##
## Help us to keep Bibliometrix free to download and use by contributing with a small donation to support our research team (https://bibliometrix.org/donate.html)
##
##
## To start with the shiny web-interface, please digit:
## biblioshiny()
2.4 The next step is to import the data. The import function is bibliometrix::convert2df(). It is important to inform as an argument the path of .bib file, the data source (dbsource) and the file format.
SCOPUS
library(bibliometrix)
file_scopus <- "https://github.com/Tai-Rocha/bibliometrix_test/raw/main/data/scopus.bib"
#file <- "https://www.bibliometrix.org/datasets/savedrecs.bib"
scopus_data <- convert2df(file = file_scopus, dbsource = "scopus", format = "bibtex")
##
## Converting your scopus collection into a bibliographic dataframe
##
##
## Warning:
## In your file, some mandatory metadata are missing. Bibliometrix functions may not work properly!
##
## Please, take a look at the vignettes:
## - 'Data Importing and Converting' (https://www.bibliometrix.org/vignettes/Data-Importing-and-Converting.html)
## - 'A brief introduction to bibliometrix' (https://www.bibliometrix.org/vignettes/Introduction_to_bibliometrix.html)
##
##
## Missing fields: CR
## Done!
##
##
## Generating affiliation field tag AU_UN from C1: Done!
WOS
file_wos <- "https://github.com/Tai-Rocha/bibliometrix_test/raw/main/data/wos.bib"
wos_data <- convert2df(file = file_wos, dbsource = "wos", format = "bibtex")
##
## Converting your wos collection into a bibliographic dataframe
##
##
## Warning:
## In your file, some mandatory metadata are missing. Bibliometrix functions may not work properly!
##
## Please, take a look at the vignettes:
## - 'Data Importing and Converting' (https://www.bibliometrix.org/vignettes/Data-Importing-and-Converting.html)
## - 'A brief introduction to bibliometrix' (https://www.bibliometrix.org/vignettes/Introduction_to_bibliometrix.html)
##
##
## Missing fields: CR
## Done!
##
##
## Generating affiliation field tag AU_UN from C1: Done!
2.5 Merge the datasets into a single dataset, which will be used to carry out the analyses.
The bibliometrix::mergeDbSources() function performs this task and using remove.duplicated = TRUE argument (for obviously remove the duplicates).
merge_all <-
bibliometrix::mergeDbSources(scopus_data, wos_data,
remove.duplicated = TRUE)
##
## 111 duplicated documents have been removed
2.6 The exported data returned different types of work, such as editorials, errata, etc.
dplyr::distinct(merge_all, DT) %>%
knitr::kable(row.names = FALSE)
| DT |
|---|
| ARTICLE |
| BOOK CHAPTER |
| REVIEW |
| CONFERENCE PAPER |
| BOOK |
| ARTICLE; PROCEEDINGS PAPER |
| PROCEEDINGS PAPER |
2.7 Filter only articles
filter_data <- merge_all %>%
filter(DT == "ARTICLE")
2.8 Now the dataset it’s ok for bibliometrics analyses. The meaning of the column names can be found in the package documentation.
dplyr::glimpse(filter_data)
## Rows: 460
## Columns: 30
## $ AU <chr> "ZHANG T;YAN Q;WANG G;ZHU J", "FISCHER S;GREET J;WALSH C;CATF…
## $ DE <chr> "NON-STRUCTURAL CARBOHYDRATES; SPROUT GROWTH; SPROUT REGENE…
## $ ID <chr> "CARBOHYDRATES; CONSERVATION; CULTIVATION; ECOSYSTEMS; FI…
## $ C1 <chr> "CAS KEY LABORATORY OF FOREST ECOLOGY AND MANAGEMENT, INSTITU…
## $ JI <chr> "ECOL. PROCESSES", "FOR. ECOL. MANAGE.", "FOR. ECOL. MANAGE."…
## $ AB <chr> "BACKGROUND: TO RESTORE SECONDARY FORESTS (MAJOR FOREST RESOU…
## $ AR <chr> "25", "119264", "119253", "119220", "119175", "119202", "1191…
## $ RP <chr> "YAN, Q.; QINGYUAN FOREST CERN, CHINA; EMAIL: QLYAN@IAE.AC.CN…
## $ DT <chr> "ARTICLE", "ARTICLE", "ARTICLE", "ARTICLE", "ARTICLE", "ARTIC…
## $ DI <chr> "10.1186/s13717-021-00300-w", "10.1016/j.foreco.2021.119264",…
## $ FU <chr> "NATIONAL NATURAL SCIENCE FOUNDATION OF CHINANATIONAL NATURAL…
## $ BN <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ SN <chr> "21921709", "03781127", "03781127", "03781127", "03781127", "…
## $ SO <chr> "ECOLOGICAL PROCESSES", "FOREST ECOLOGY AND MANAGEMENT", "FOR…
## $ LA <chr> "ENGLISH", "ENGLISH", "ENGLISH", "ENGLISH", "ENGLISH", "ENGLI…
## $ TC <dbl> 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 2, 0, 2, 1, 0, 0…
## $ PN <chr> "1", NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "10", "5", N…
## $ PP <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "…
## $ PU <chr> "SPRINGER SCIENCE AND BUSINESS MEDIA DEUTSCHLAND GMBH", "ELSE…
## $ DB <chr> "ISI", "ISI", "ISI", "ISI", "ISI", "ISI", "ISI", "ISI", "ISI"…
## $ TI <chr> "THE EFFECTS OF STUMP SIZE AND WITHINGAP POSITION ON SPROUT N…
## $ VL <chr> "10", "493", "493", "492", "491", "491", "491", "490", "489",…
## $ PY <dbl> 2021, 2021, 2021, 2021, 2021, 2021, 2021, 2021, 2021, 2021, 2…
## $ FX <chr> "THIS RESEARCH WAS SUPPORTED BY GRANTS FROM THE STRATEGIC LEA…
## $ CR <chr> "none", "none", "none", "none", "none", "none", "none", "none…
## $ AU_UN <chr> "INSTITUTE OF APPLIED ECOLOGY;CHINESE ACADEMY OF SCIENCES;CLE…
## $ AU1_UN <chr> "NOTREPORTED;QINGYUAN FOREST CERN;NOTREPORTED", "FISCHER;SCHO…
## $ AU_UN_NR <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ SR_FULL <chr> "ZHANG T, 2021, ECOL PROCESSES", "FISCHER S, 2021, FOR ECOL M…
## $ SR <chr> "ZHANG T, 2021, ECOL PROCESSES", "FISCHER S, 2021, FOR ECOL M…
3. Bibliometrix functions for analyzing data
3.1 The bibliometrix::biblioAnalysis() function provides a summary of the dataset. I won’t present the result here once it’s too long, but you can see an example in thistutorial.
summary <- bibliometrix::biblioAnalysis(filter_data)
The plot() function builds graphics when we offer the result of the bibliometrix::biblioAnalysis() function. Several standardized graphics are automatically generated:
graphics_bibliometrix <- plot(summary)
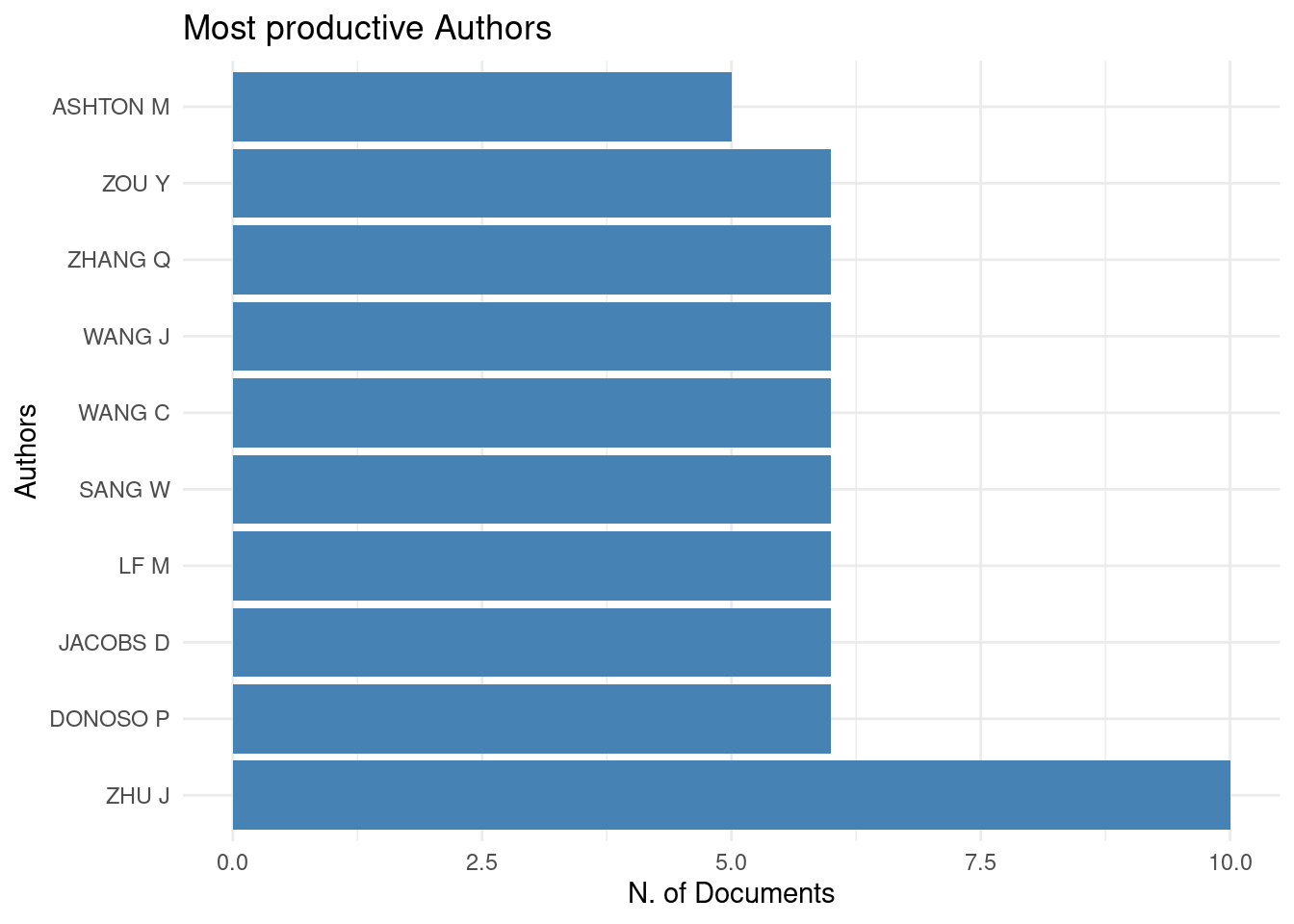
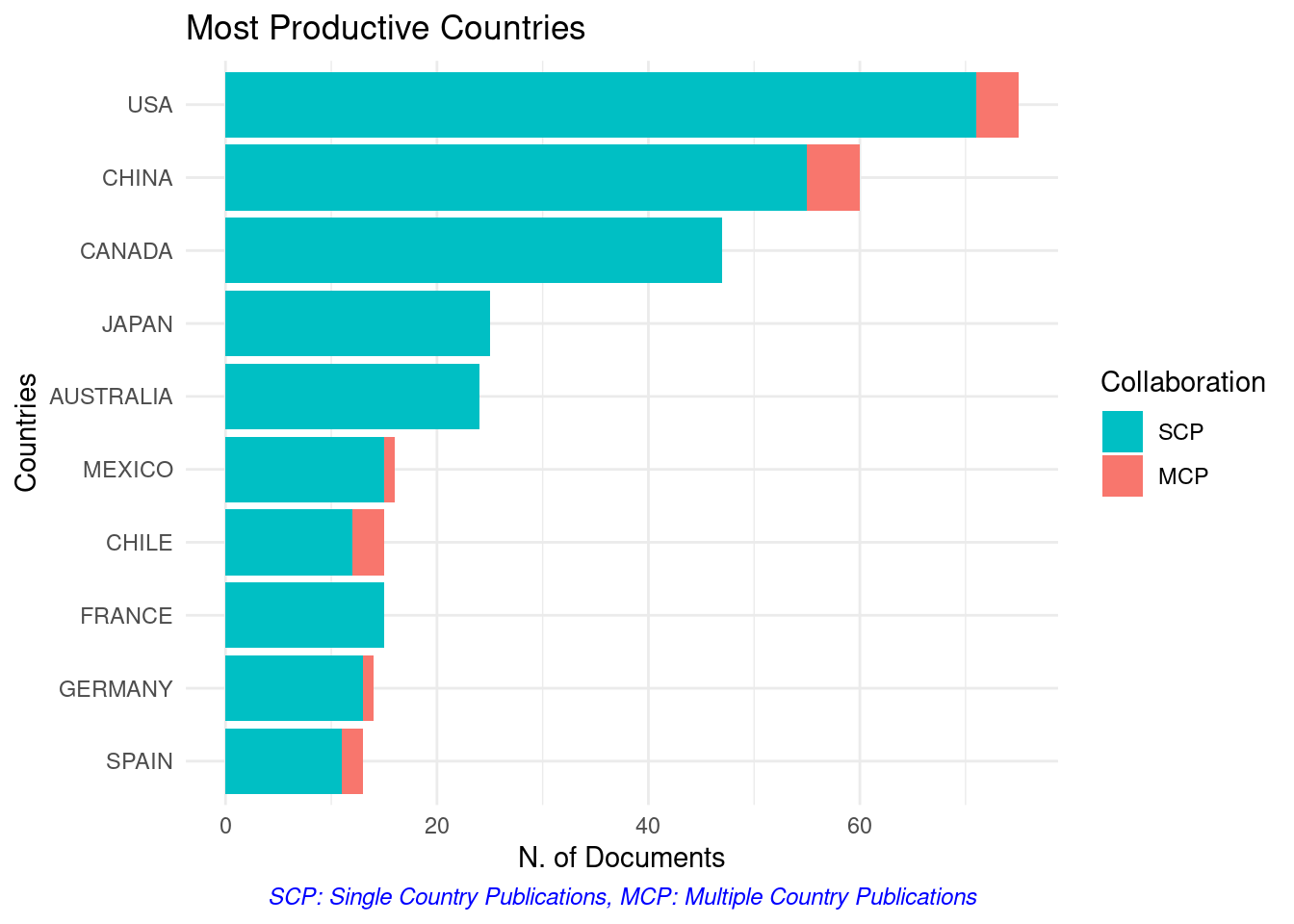

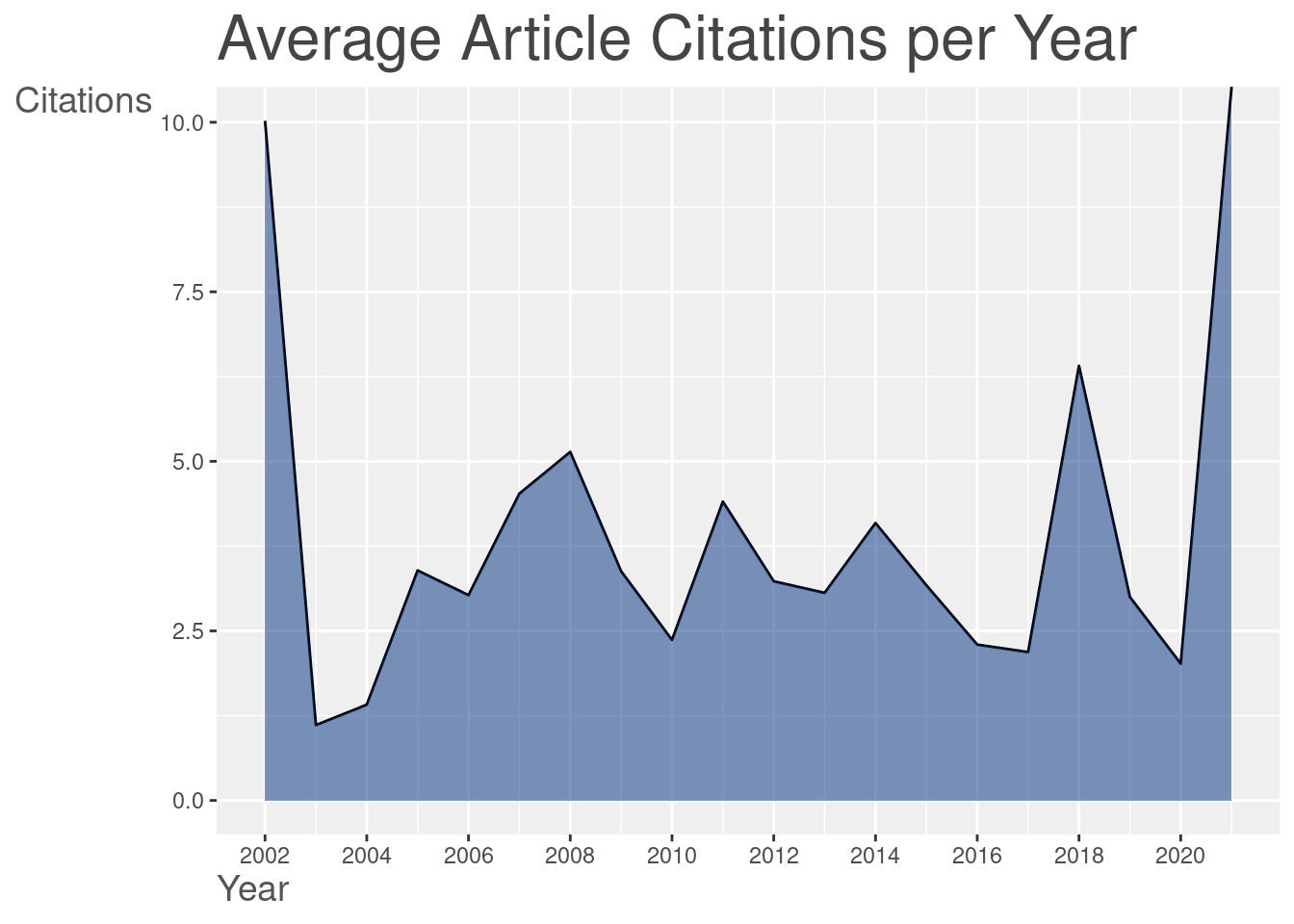
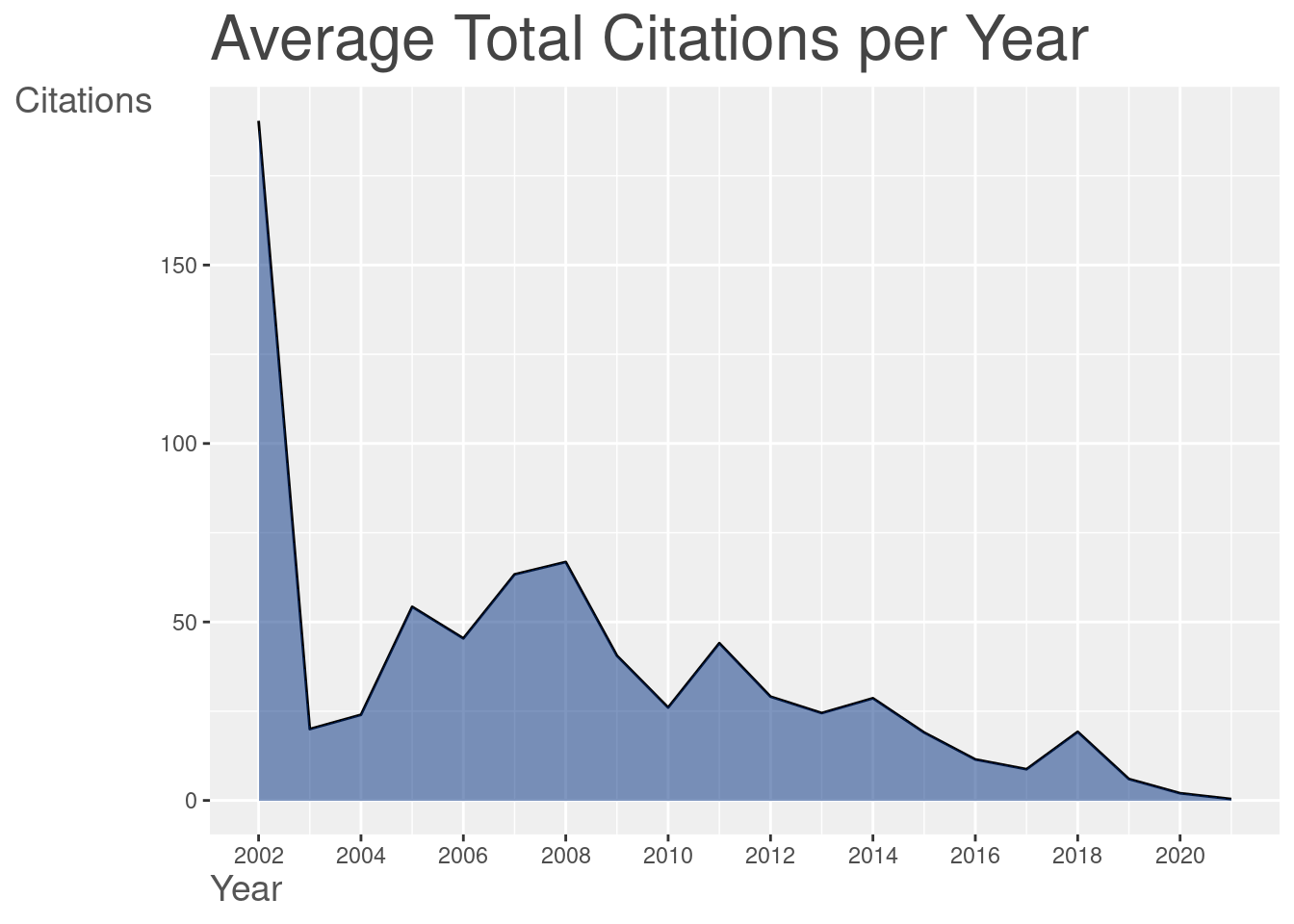
4 References
Aria, Massimo, and Corrado Cuccurullo. 2017. “Bibliometrix: An r-Tool for Comprehensive Science Mapping Analysis.” Journal of Informetrics 11 (4): 959–75. https://doi.org/10.1016/j.joi.2017.08.007.
Bibliometrix: Comprehensive Science Mapping Analysis (2021). https://CRAN.R-project.org/package=bibliometrix .
© 2021 GitHub, Inc. Terms Privacy Security Status Docs Contact GitHub Pricing API Training Blog About
License

Content is available under the Creative CommonsAttribution-ShareAlike (CC BY-SA) license. You can share and adapt it, but you must attribute the credits to the authors, adding a link to the original content. , and your sharing must also have this same type of license.
More info: Creative Commons
- Posted on:
- 11/12/2021
- Length:
- 6 minute read, 1164 words
- See Also: